Tertiary (3˚) Structure
Proteins are abundant in all organisms and are fundamental to life. The diversity of protein structure underlies the very large range of their functions: enzymes (biological catalysts), storage, transport, messengers, antibodies, regulation, and structural proteins.
Proteins are linear heteropolymers of fixed length; i.e. a single type of protein always has the same number and composition of AAs, but different proteins may have 100 to more than 1000 AAs. There is therefore a great diversity of possible protein sequences. The linear chains fold into specific three-dimensional conformations, which are determined by the sequence of amino acids and therefore are also extremely diverse, ranging from completely fibrous to globular. Covalent disulfide bonds can be introduced between cysteine residues placed in close proximity in 3D space -- this provides rigidity for the resulting 3D structure. Ribbon diagrams like the one shown here are a common way to visualize proteins.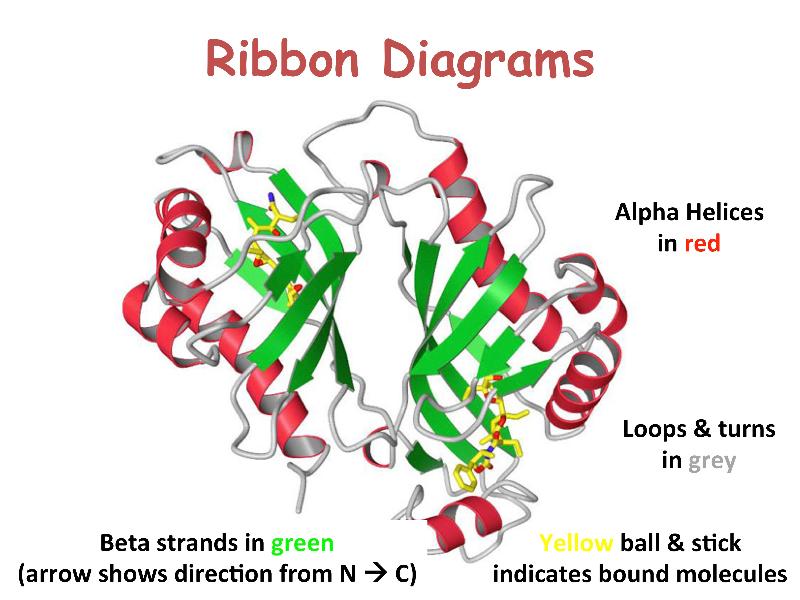
Protein structures can be determined to an atomic level by X-ray diffraction and neutron-diffraction studies of crystallized proteins, and more recently by nuclear magnetic resonance (NMR) spectroscopy of proteins in solution. The structures of many proteins, however, remain undetermined.
To view an example of tertiary structure in KiNG, click here. This is ribonuclease A, an enzyme responsible for the degradation of RNA. The image depicts all atoms of one half of the molecule (cyan for side chains, brown for hydrogen atoms) and just main chain and side chains for the other half. The alternate view shows main-chain atoms and H-bonds (purple). Click "Animate" to cycle between the views.
Although hydrogens constitute about half the atoms in a protein, they are seldom shown explicitly because they are hard to detect with x-ray crystallography (due to low electron density) and they very much complicate the picture. This ribonuclease image is a joint x-ray/neutron diffraction structure, for which hydrogens are always included. Even without H atoms, an all-atom view is too crowded to be very useful but is a good way to appreciate where simplified versions start from.
Some text adapted from: Kinemage Supplement to Branden & Tooze "Introduction to Protein Structure", Chapter 2 - MOTIFS OF PROTEIN STRUCTURE, Jane S. and David C. Richardson.