Sequence Alignment
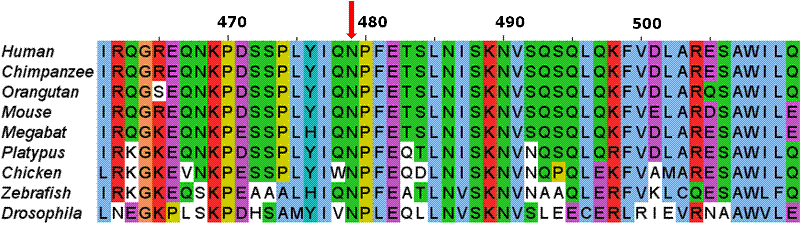 The first molecular sequences available to scientists were protein. To unravel evolutionary relationships a scientist would align these sequences by hand looking for regions of similarity. Today computers compare DNA, RNA and/or protein sequences using very elegant algorithms. We believe regions of similarity represent a shared common ancestor, a shared homology. The relationship between the aligned sequences is often described with the terms "percent homology" or "sequence similarity". An example of such an alignment is displayed to the right.
The first molecular sequences available to scientists were protein. To unravel evolutionary relationships a scientist would align these sequences by hand looking for regions of similarity. Today computers compare DNA, RNA and/or protein sequences using very elegant algorithms. We believe regions of similarity represent a shared common ancestor, a shared homology. The relationship between the aligned sequences is often described with the terms "percent homology" or "sequence similarity". An example of such an alignment is displayed to the right.
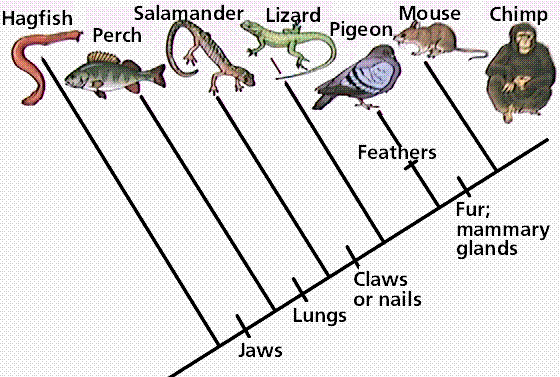
Pioneered by the German entomologist Willi Hennig in his work in the 1960s, cladistics is an approach to biological classification in which items are grouped together based on whether or not they have one or more shared unique characteristics that come from the group's last common ancestor and are not present in more distant ancestors. Therefore, members of the same group are thought to share a common history and are considered to be more closely related. In the simple claudogram (tree) to the left you can see the characteristics that they used to separate the branches (species) along the root.
As more proteins were sequenced it became possible to compare the same protein between different species. An example of such an analysis is shown to the left, where change over evolutionary time of fibrinopeptides (peptides which keep fibrinogen from clotting), hemoglobins, cytochrome c (used in the electron transport chain), and histone H4s (used to package DNA up in the nuclei) are compared. Why aren't these proteins all changing at the same rate?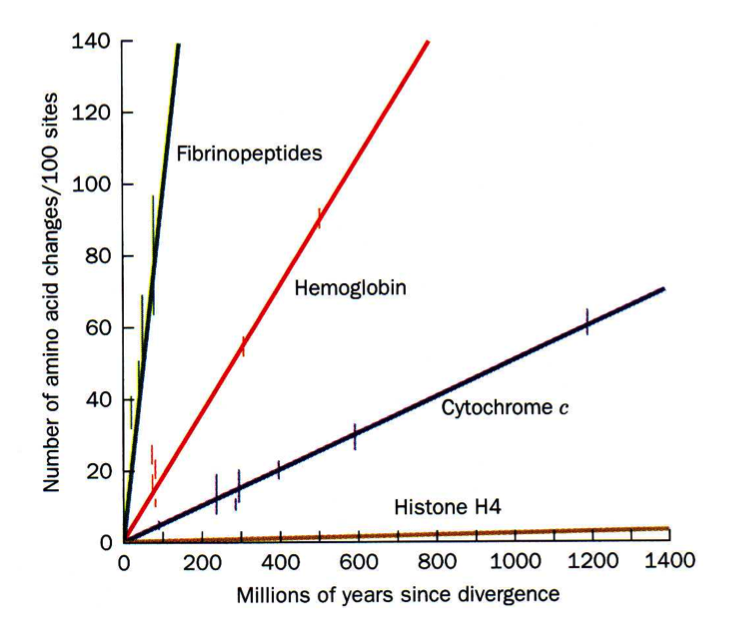
When these data were compared to 3D structures it became apparent that the AAs that changed most frequently were those located on the surfaces of proteins (what type of AAs would you expect these to be?) and those that rarely changed were located on the interior (what type of AAs would you expect these to be?).