Protein Architecture
Foundations of Clinical Sciences
Initially biochemists believed that each protein would have a unique architecture, but that idea quickly faded as we started to obtain 3˚ structures. We now believe protein structural diversity encompasses a finite set of architectural designs. The current Structural Genomics Project supported by NIH attempts to determine the structure of every protein encoded by the genome. Since protein structure is closely linked to protein function, the structural genomics has the potential to inform knowledge of protein function. In addition to elucidating protein functions, structural genomics can be used to identify novel protein folds and potential targets for drug discovery.
In this module we will examine patterns and architectural elements shared by different proteins.
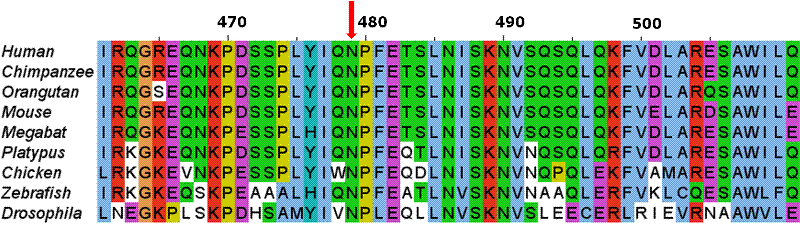 The first molecular sequences available to scientists were protein. To unravel evolutionary relationships a scientist would align these sequences by hand looking for regions of similarity. Today computers compare DNA, RNA and/or protein sequences using very elegant algorithms. We believe regions of similarity represent a shared common ancestor, a shared homology. The relationship between the aligned sequences is often described with the terms "percent homology" or "sequence similarity". An example of such an alignment is displayed to the right.
The first molecular sequences available to scientists were protein. To unravel evolutionary relationships a scientist would align these sequences by hand looking for regions of similarity. Today computers compare DNA, RNA and/or protein sequences using very elegant algorithms. We believe regions of similarity represent a shared common ancestor, a shared homology. The relationship between the aligned sequences is often described with the terms "percent homology" or "sequence similarity". An example of such an alignment is displayed to the right.
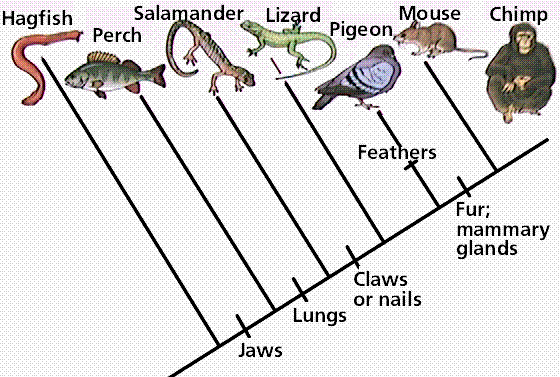
Pioneered by the German entomologist Willi Hennig in his work in the 1960s, cladistics is an approach to biological classification in which items are grouped together based on whether or not they have one or more shared unique characteristics that come from the group's last common ancestor and are not present in more distant ancestors. Therefore, members of the same group are thought to share a common history and are considered to be more closely related. In the simple claudogram (tree) to the left you can see the characteristics that they used to separate the branches (species) along the root.
As more proteins were sequenced it became possible to compare the same protein between different species. An example of such an analysis is shown to the left, where change over evolutionary time of fibrinopeptides (peptides which keep fibrinogen from clotting), hemoglobins, cytochrome c (used in the electron transport chain), and histone H4s (used to package DNA up in the nuclei) are compared. Why aren't these proteins all changing at the same rate?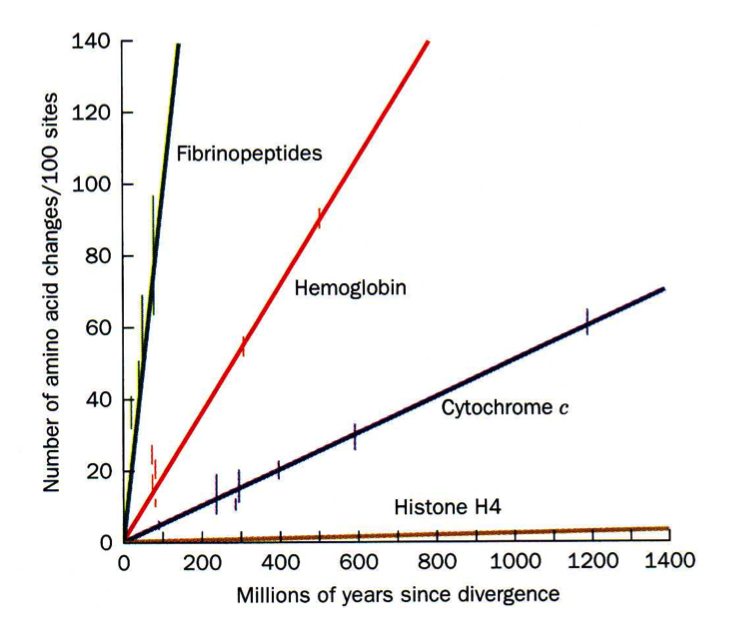
When these data were compared to 3D structures it became apparent that the AAs that changed most frequently were those located on the surfaces of proteins (what type of AAs would you expect these to be?) and those that rarely changed were located on the interior (what type of AAs would you expect these to be?).
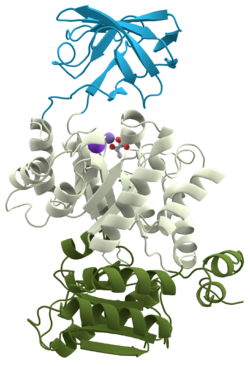 Some of the first proteins utilized in the biochemical laboratory were proteases -- proteins that cleave other proteins. It was frequently observed, however, that a protein would be digested to one or more fragments that could not be digested further except by introducing high temperature or denaturing agents. In 1981 these were named domains and they are now defined as " within a single subunit [polypeptide chain], a contiguous portion which folds into a compact, local semi-independent unit ...". As mentioned, domains are separable by proteases and have frequently been found to have a specific function (binding, digesting, etc). A domain can evolve, function, and maintain its structure relatively independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded, thus they can easily be expressed in a laboratory setting. Typically we find globular proteins to be organized into one or more domains. The ribbon structure (shown to the left) is for pyruvate kinase, a protein with three domains (each colored differently).
Some of the first proteins utilized in the biochemical laboratory were proteases -- proteins that cleave other proteins. It was frequently observed, however, that a protein would be digested to one or more fragments that could not be digested further except by introducing high temperature or denaturing agents. In 1981 these were named domains and they are now defined as " within a single subunit [polypeptide chain], a contiguous portion which folds into a compact, local semi-independent unit ...". As mentioned, domains are separable by proteases and have frequently been found to have a specific function (binding, digesting, etc). A domain can evolve, function, and maintain its structure relatively independently of the rest of the protein chain. Each domain forms a compact three-dimensional structure and often can be independently stable and folded, thus they can easily be expressed in a laboratory setting. Typically we find globular proteins to be organized into one or more domains. The ribbon structure (shown to the left) is for pyruvate kinase, a protein with three domains (each colored differently).
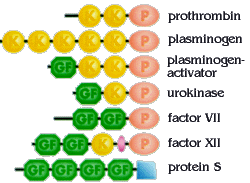
During evolution, DNA segments coding for individual domains of proteins have been duplicated and rearranged. This has given rise to families of related proteins that share similar domains. Blood coagulation factors represent such a family, with members containing similar domains in various combinations and numbers. P=protease domain, GF=growth factor domain, K="kringle"-domain. Coagulation proteins are a great example of proteins being modular.
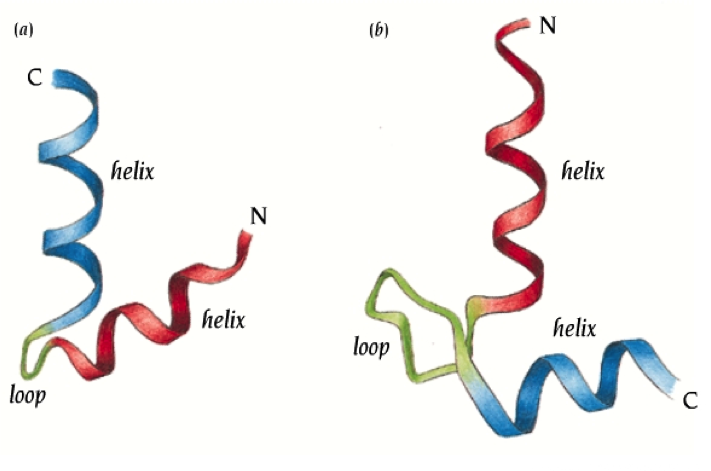
Motifs were discovered after many 3D structures were available for examination. A motif is a simple geometric arrangement of one or more secondary structural elements -- for example helix-turn-helix (see image) used in DNA binding -- that occurs in many different proteins. It falls somewhere between secondary and tertiary structure in terms of level of organizaiton. Unlike domains, motifs are incapable of independent folding. A domain may contain zero, one, two, etc motifs within its structure.